AI: Language model enables protein evolution in fast-forward mode
EvolutionaryScale can design proteins with desired properties according to prompts by simultaneously evolving sequence, structure and function.
(Image: EvolutionaryScale)
A new artificial intelligence (AI) called ESM3 can design proteins that would have taken hundreds of millions of years to evolve. This is what EvolutionaryScale, a US start-up founded by former Meta employees, writes in a preprint on the BioRxiv platform.
Their generative masked language model is not only one of the largest biological AIs. It is also the first model that can simultaneously work out the amino acid sequence, the 3D structure and also the function of a protein with a desired ability after prompts have been entered. Future areas of application range from drug development and materials science to proteins for storing carbon dioxide.
The 3D structure of proteins is one of the most important pieces of information in biology and pharmacy. Proteins are like tiny bio-machines that shape our bodies and keep them running, for example as building material in muscles, hair and nails, as hormones and as antibodies. Knowing the shape of proteins helps to elucidate their biological function in the body, determine their effectiveness as drugs and test their suitability as drug targets.
Videos by heise
Many life-saving drugs are proteins, for example insulin for diabetics as well as artificial antibodies against cancer and also against severe respiratory infections caused by RSV (respiratory syncytial virus). In medicine in particular, however, the aim is often to synthesize completely new proteins with desired properties instead of laboriously searching for them.
Masking across three levels
For this new synthesis, EvolutionaryScale uses a masked language model for ESM3. The model can not only infer missing (masked) information or tokens in one category by checking the context in both directions as in a text, but also across three categories. ESM3 was trained with protein data in which all three categories were known – a total of 2.8 billion amino acid sequences, 236 million protein structures and 539 million protein functions.
The developers created a separate alphabet for the sequence, 3D structure and function of the proteins and developed a method for describing each 3D structure as a sequence of letters. If changing parts of the information were then masked in all three categories, the language model learned to understand not only the context within the individual levels, but also between the levels.
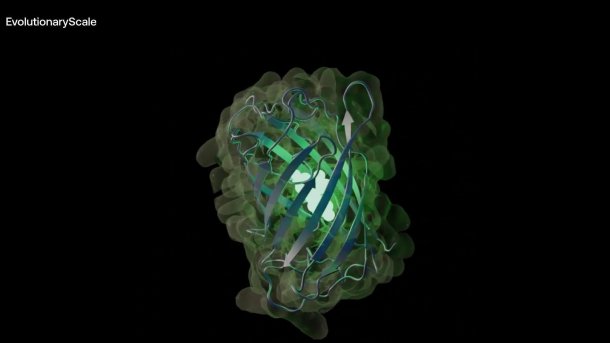
Synthetic variants of a green fluorescent protein
To demonstrate the performance of ESM3, the start-up had the language model design synthetic variants of the green fluorescent protein (GFP), which also glow well. GFP is a natural protein that, in different variants, makes marine animals such as jellyfish and corals glow and is one of the most important molecules in molecular biology research. Its discovery was rewarded with the Nobel Prize in Chemistry in 2008. GFP can be used, for example, to mark molecules in living cells in order to observe biological processes that would otherwise be inaccessible, such as the development of nerve cells in the brain or how cancer cells spread.
The best artificial GFP variant "esmGFP" from ESM3 now shone as brightly as a natural GFP variant from the training data. The good luminosity of the new design was based on a gene blueprint that was surprisingly close to the blueprint of the underlying natural template, namely only 58 percent. "Based on the diversification rate of GFPs in nature, we estimate that the generation of a new fluorescent protein corresponds to the simulation of over 500 million years of evolution," write the ESM3 developers.
Alex Rives, EvolutionaryScale's lead scientist, had previously worked with his colleagues on earlier versions of the ESM model at Meta. However, after Meta discontinued its work in this area last year, the developers opted for independent development. With success: at the same time as announcing the new fluorescent protein, the start-up also announced an investment injection of 142 million dollars to bring compounds into use.
At the same time, EvolutionaryScale also released a smaller, open-access version for scientists that does not contain the full functionality. Researchers, such as Martin Pacesa from the Swiss Federal Institute of Technology in Lausanne, are looking forward to testing the language model extensively. However, the structural biologist also warned the journal "Nature" that academic groups would not be able to replicate their own full version, as this would require enormous computing resources.
(emw)